Cancer is a serious disease threatening to human health; its incidence is on the increase every year. The research group of the molecular cell biology of cancer focuses mainly on the expression and epigenetic regulatory mechanism of important genes which are related with the process of tumor genesis, development and metastasis. In addition, the group also pays attention to the alteration i n signal transduction pathway of cancer cells, the structure characteristics and its mechanism of cancer cells, the glycan structure of cancer cell surface and cancer metabolomics. The goal of the group is to elucidate the molecular mechanism of cancer genesis, development and metastasis, and explore new ways for early diagnosis and treatment of cancer by the development of transformational medicine technology.
Introduction:
The staff consists of 1 professor, 4 associated professors and 3 lecturers i n the group. In addition, 2 postdoc, 8 PhD students and 10 master students are engaged in. Most of staff member have overseas study or research experience, and have publishing records with academic value i n key domestic and international professional journals. The main group members include Li Yu, Shi Shuliang, He Jie, Tian Weiming, Nie Huan, Han fang, Han Fengtong and Zhang Hao.
Recent years, the group has been granted more than 4.9 million and 20 projects including 7 National Natural Science Funds Fund(NSFC), 3 National High-tech Program of China (863 Program), 2 Major national scientific research plan (973 Program), 3 National Lab projects and 7 other provincial and ministerial level projects. Over 30 SCI indexed research papers have been published totally.
Teaching courses:
The teachers of the group undertake the teaching task on the PhD, master and undergraduate students including biochemistry, cell biology, molecular biology, immunology, molecular genetics, molecular cell biology, technique of immunology, molecular cell biology of cancer, cell engineering, gene engineering, and structure and function of biomacromolecules. Furthermore the lab is open for all students and provides the technical support to innovation research.
Student recruitment:
The group could enroll Master of Science on the direction of cell biology, biochemistry and molecular biology i n biology discipline, and Doctor of Science on the direction of the molecular cell biology of cancer i n biomedical engineering discipline. Also, could enroll post-doctor.
International communications:
We have collaborated closely with the Harvard University, School of Medicine, Texas A and M University i n the area of cancer biology, and Cleveland State University i n the area of Glycomics. The collaboration includes cooperating training of graduate students, junior scientist training and collaborative research, and contract professor engagement.
The main research fields:
1) The study on the function and regulation of cancer related gene and protein
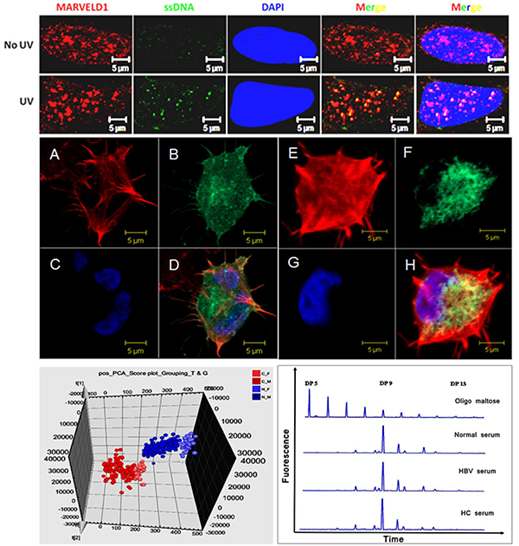
The main research fields are functional identification and expression regulation of new genes that are closely related with carcinogenesis. In the aspect of expression regulation, we focus on the epigenetic regulation mechanism including histone methylation, histone acetylation, DNA methylation and related regulation. Meanwhile, the group also conducts researches on the regulation mechanism and the downstream target of cancer related miRNA. On the other hand, we aim to clarify some molecular and cellular biology issues such as cell cycle and cell proliferation, apoptosis, DNA damage and repair, signal transduction, cytoskeleton dynamic assembly, cell-matrix interactions and nuclear protein interactions which are related to carcinogenesis. Now we are interested i n a series of cancer related genes including MARVELD1, c1orf109, SALL4, BI-1, BRI-1, AIF and some interaction molecules associated with histone methylation.
2) The research on the signal transduction of cancer cell
Biological phenotype of cancer is originated from the disturbance of cell signal transduction system. To investigate the mechanisms of the aberrant signal transduction involved i n cell proliferation, apoptosis, adhesion, migration and invasion, the group focuses on the meaning of the activation and regulation of Ras family small G proteins including Ras, Rap, Rab ,Rho, Ran and downstream signal pathway i n various cancer type and i n different pathology phage. We are intensively investigating the regulatory mechanism of abnormal signal transduction of cancer, including signaling pathways inside the cells, and on cell-cell and cell-matrix contacts during the process of the carcinogenesis, along with the molecular events leading to specific biological phenotype of cancer. To reveal the physiological and biochemical change of cancer, which underlies the molecular mechanism of carcinogenesis, cell line model, animal model and clinic tissue sample were used, and the combined techniques including protein-protein interaction, protein activation, RNAi, cell immuno-fluorescence and live cell imaging assays were included as the methodology.
3) The study of the cancer glycomics and biomarker
Taking the third biomacromolecules glycan as the research object, the specific glycan changes i n serum, cell lines and tissue from the patients of breast cancer, hepatocarcinomas and pancreatic cancer were studied, based on the improved high throughput analytical technique of N-glycan. In addition, the relation between the change of the glycan and the function of glycoprotein was also investigated. The ultimate aims are to build a rapid, convenient and noninvasive blood detection technique, by exploring the sensitive and specific cancer biomarker, and provide new cues to tackle down the target for cancer therapy.
4) The study of the cancer metabolomics and biomarker
On the basis of developing a research platform of metabolomics with the UPLC-MS/MS, the group has studied the metabolomics of serum, urine and tissue sample of various cancers. The goal is to find the micro-molecular metabolomic biomarker of cancer through the multi-element statistical analysis and to develop the clinical diagnostic models for early diagnosis of associated cancer.
Key Publications in recent years:
1.Ming Shi, Shan Wang, Yuanfei Yao, Yiqun Li, Hao Zhang, Fang Han, Huan Nie, Jie Su, Zeyu Wang, Lei Yue, Jingyan Cao, Yu Li*. Biological and clinical significance of epigenetic silencing of MARVELD1 gene in lung cancer. Scientific Reports. 2014; 4: 7545, doi:10.1038/srep07545.
2.Jiang Q, Wang J, Wu X, Ma R, Zhang T, Jin S, Han Z, Tan R, Peng J, Liu G, Li Y, Wang Y. LncRNA2Target: a database for differentially expressed genes after lncRNA knockdown or overexpression. Nucleic Acids Res. 2014 Nov 15. pii: gku1173.
3.Xu S, Wu H, Nie H, Yue L, Jiang H, Xiao S, Li Y*. AIF downregulation and its interaction with STK3 in renal cell carcinoma. PLoS One. 2014 Jul 3; 9(7):e100824. doi: 10.1371/journal.pone.0100824.
4.Wei L, Liu C, Kang L, Liu Y, Shi S, Wu Q, Li Y*. Experimental study on effect of simulated microgravity on structural chromosome instability of human peripheral blood lymphocytes. PLoS One. 2014 Jun 25; 9(6):e100595. doi: 10.1371/journal.pone.0100595.
5.Jiang Q, Wang J, Wang Y, Ma R, Wu X, Li Y. TF2LncRNA: identifying common transcription factors for a list of lncRNA genes from ChIP-Seq data. Biomed Res Int. 2014; 2014:317642. doi: 10.1155/2014/317642.
6.Shi M, Yao Y, Han F, Li Y, Li Y*. MAP1S controls breast cancer cell TLR5 signaling pathway and promotes TLR5 signaling-based tumor suppression. PLoS One. 2014 Jan 23; 9(1):e86839. doi: 10.1371/journal.pone.0086839.
7.Jiang Q, Wang G, Jin S, Li Y, Wang Y. Predicting human microRNA-disease associations based on support vector machine. Int J Data Min Bioinform. 2013;8(3):282-93.
8.Narla SN, Pinnamaneni P, Nie H, Li Y, Sun XL. BSA-boronic acid conjugate as lectin mimetics. Biochem Biophys Res Commun. 2014 Jan 10;443(2):562-7. doi: 10.1016/j.bbrc.2013.12.006.
9.Wei L, Diao Y, Qi J, Khokhlov A, Feng H, Yan X, Li Y*. Effect of change in spindle structure on proliferation inhibition of osteosarcoma cells and osteoblast under simulated microgravity during incubation in rotating bioreactor. PLoS One. 2013 Oct 7;8(10):e76710. doi: 10.1371/journal.pone.0076710.
10.Shan Wang, Jianran Hu, Yuanfei Yao, Ming Shi, Lei Yue, Fang Han, Hao Zhang, Jie He, Shanshan Liu, Yu Li*. MARVELD1 regulates integrin β1-mediated cell adhesion and actin organization via inhibiting its pre-mRNA processing. Int J Biochem Cell Biol. 2013; doi.org/doi:10.1016/j.biocel.2013.09.006
11.Liu X, Nie H, Zhang Y, Yao Y, Maitikabili A, Qu Y, Shi S, Chen C, Li Yu*. Cell surface-specific N-glycan profiling i n breast cancer. PLoS One. 2013; 8(8):e72704. doi: 10.1371/journal.pone.0072704.
12.He J, Zhang W, Zhou Q, Zhao T, Song Y, Chai L, Li Yu*. Low-expression of microRNA-107 inhibits cell apoptosis i n glioma by upregulation of SALL4. Int J Biochem Cell Biol. 2013; 45(9):1962-73. doi: 10.1016/j.biocel.2013.06.008.
13.Ni S, Hu J, Duan Y, Shi S, Li R, Wu H, Qu Y, Li Yu*. Down expression of LRP1B promotes cell migration via RhoA/Cdc42 pathway and actin cytoskeleton remodeling i n renal cell cancer. Cancer Sci. 2013; 104(7):817-25. doi: 10.1111/cas.12157.
14.Ling S, Birnbaum Y, Nanhwan MK, Thomas B, Bajaj M, Li Y, Li Y, Ye Y. Dickkopf-1 (DKK1) phosphatase and tensin homolog on chromosome 10 (PTEN) crosstalk via microRNA interference i n the diabetic heart. Basic Res Cardiol. 2013; 108(3):352. doi: 10.1007/s00395-013-0352-2.
15.Yan Hongji, He Yue, Yin Yanbin, Wang Xiumei, Xiongbiao Chen, Song Zhang, Cui Fuzhai, Yu Li, Yongzhan Nie, Weiming Tian. Self-assembled monolayers with different chemical group substrates for the study of MCF-7 breast cancer cell line behavior. Biomedical Materials, 2013; 8(3):035008. doi: 10.1088/1748-6041 /8/3/035008.
16.Xiuping Bai, Rui Fang, Song Zhang, Xinli Shi, Zeli Wang, Xiongbiao Chen, Jing Yang, Xiaolu Hou, Yongzhan Nie, Yu Li*, and Weiming Tian*. Self-cross-linkable hydrogels composed of partially oxidized alginate and gelatin for myocardial infarction repair. Journal of Bioactive and Compatible Polymers, first published on February 5, 2013 as doi:10.1177/0883911512473230
17.Qinghua Jiang, Guohua Wang, Shuilin Jin, Yu Li, Yadong Wang. Predicting human microRNA-disease associations based on support vector machine. Int. J. Data Mining and Bioinformatics 2013; 8(3), 282-293.
18.Wang haochang, Li Yu*. Co-decision Matrix Framework for Name Entity Recognition in Biomedical Text. International Journal of Data Mining and Bioinformatics. 2013.
19.Wang haochang, Li Yu*. Transductive Support Vector Machines and Active Learning for Extracting Protein-Protein Interactions. Journal of Investigative Medicine. 2013; 61(4)
20.Lijun Wei, Fang Han, Lei Yue, Hongxia Zheng, Dan Yu, Xiaohuan Ma, Huifang Cheng, Yu Li *. Synergistic Effects of Incubation i n Rotating Bioreactors and Cumulative Low Dose 60Co γ-ray Irradiation on Human Immortal Lymphoblastoid Cells. Microgravity Science and Technology: 2012, 24 (5):335-344
21.Huan Nie, Yu Li* and Xue-Long Sun*. Recent advances i n sialic acid-focused glycomics. Journal of Proteomics. 2012; 75(11):3098-112.
22.Shan-shan Liu, Hong-xia Zheng, Hua-dong Jiang, Jie He, Yang Yu, You-peng Qu, Lei Yue, Yao Zhang, Yu Li*. Identification and characterization of a novel gene, c1orf109, encoding a CK2 substrate that is involved i n cancer cell proliferation. Journal of biomedical science 2012; 19:49.
23.Youtao Yu, Yubao Zhang, Jianran Hu, Hao Zhang , Shan Wang, Fang Han, Lei Yue, Youpeng Qu, Yao Zhang, Hongjian Liang, Huan Nie, Yu Li*. MARVELD1 inhibited cell proliferation and enhance chemosensitivity via increasing expression of p53 and p16 in hepatocellular carcinoma. Cancer Science. 2012;103(4):716-22.
24.Satya Nandana Narla, Huan Nie, Yu Li*, Xue-Long Sun*. Recent Advances in the Synthesis and Biomedical Applications of Chain-End Functionalized Glycopolymers. Journal of Carbohydrate Chemistry, 2012; 31(2):67-92
25.Wenying Mu, Shanguang Chen, Fengyuan Zhuang, Yinghui Li, Yu Li. Quantification of the distribution of blood flow pressure with postures. J Biomedical Science and Engineering( JbiSE), 2012; 5, 113-119 doi:10.4236/jbise.2012.53015
26.Hong-xia Zheng, Shan-shan Liu, Wei-ming Tian, Hong-ji Yan, Yao Zhang, Yu Li*. A three-dimensional i n vitro culture model for primary neonatal rat ventricular myocytes. Current applied physics, 2012; 12(3):826-833.
27.Zheng Hongxia, Tian Weiming, Yue Lei, Zhang Yao, Yu Li*. Rotary culture promotes the proliferation of MCF-7 cells encapsulated i n three-dimensional collagen-alginate hydrogels via activation of ERK1/2-MAPK pathway, Biomedical materials, 2012; 7(1):015003.
28.Zheng Hongxia, Tian Weiming, Yan Hongji, Jiang Huadong, Liu Shan-shan, Yue Lei, Zhang Yao, Han Fengtong, Yu Li*. Expression of estrogen receptor α i n human breast cancer cells regulated mitochondrial oxidative stress under simulated microgravity. Advances i n Space Research, 2012; 49(10):1432-1440.
29.Liu SS, Chen XM, Zheng HX, Shi SL, Yu Li*. Knockdown of Rab5a expression decreases cancer cell motility and invasion through integrin-mediated signaling pathway. J Biomed Sci. 2011; 18:58.
30.X. P. Bai, H. X. Zheng, R. Fang, T. R. Wang, X. L. Hou, Yu Li, X. B. Chen, W. M. Tian*. Fabrication of engineered heart tissue grafts from alginate/collagen barium composite microbeads. Biomedical materials. 2011; 6(4):045002.
31.Liu SS, Chen XM, Zheng HX, Shi SL, Yu Li*. Knockdown of Rab5a expression decreases cancer cell motility and invasion through integrin-mediated signaling pathway. J Biomed Sci. 2011; 18:58.
32.Fanli Zeng, Yanyan Tian, Shuliang Shi, Qiong Wu, Lei Yue, Yao Zhang, Yu Li*. Identification of mouse MARVELD1 as a candidate microtubule-associated protein that inhibites cell-cycle progression and migration. Molecules and Cells. 2011; 31(3):267-274.
33.Ning Ding, Huan Nie, Xuemei Sun, Wei Sun, Youpeng Qu, Xia Liu, Yuanfei Yao, Xue Liang, Cuiying Chitty Chen, Yu Li*. Human Serum N-Glycan Profiles Are Age and Sex Dependent. Age and Aging, 2011, 40(5):568-75. doi: 10.1093/ageing/afr084
34.Hongxia Zheng, Weiming Tian, Lei Yue, Lijun Wei, Yu Li*. (2010) Simulated weightlessness leads to structural aberrant spindle apparatus and cell cycle arrest i n NIH3T3 fibroblast cells. Space Medicine & Medical Engineering, 23(6)416-418
35.Shan Wang,Yu Li*, Fang Han, Jianran Hu, Lei Yue, Youtao Yu, Yubao Zhang, Jie He, Hongxia Zheng, Shuliang Shi, Xiaowei Fu, Hongjin Wu. Identification and characterization of MARVELD1, a novel nuclear protein that is downregulated i n multiple cancers and silenced by DNA methylation. Cancer Letters. 2009; 282:77-86.
36.Fangli Liu, Yu Li, Yang Yu, Songbin Fu and Pu Li. Cloning of Novel Tumor Metastasis-Related Genes from the Highly Metastatic Human Lung Adenocarcinoma Cell Line Anip973. Journal of Genetics and Genomics. 2007; 34: 189-195
37.Li Yu, Qing Chang, Brian P. Rubin, Christopher D.M. Fletcher, Thomas W. Morgan, Steven J. Mentzer, David J.Sugarbaker, Jonathan A. Fletcher, and Sheng Xiao. Insulin Receptor Activation i n Solitary Fibrous Tumors. J. of Pathology. 2007; 211:550-554.
38.Huang JM, Zhao YL, Li Yu, Fletcher JA, Xiao S. Genomic and functional evidence for an ARID1A tumor suppressor role. Genes Chromosomes Cancer. 2007; 46:745-750.
39.Li Yu, Jianmin Huang, Yan-Ling Zhao, Ji He, Wei Wang, Kay E. Davies, Vânia Nosé, and Sheng Xiao. UTRN on chromosome 6q24 is mutated i n multiple tumors. Oncogene 2007; 26: 6220-6228.